Surface Markers: Definition and Significance
Surface markers are protein, carbohydrate, or lipid molecules embedded in the cell membrane. Each type of cell has a specific set of surface markers that makes it unique. Surface markers include molecules that are able to bind various other molecules or cells, or antibodies directed against them. Antibodies recognizing similar antigens are grouped into clusters, and are known as clusters of differentiation (CD).
Function of Cell Surface Markers
1. Cell Type Identification and Classification
Think of surface markers like fingerprints. Each cell type possesses a distinct fingerprint of markers, allowing their identification and classification. This is crucial for:
- Immune System: B cells use specific surface markers on pathogens to identify and target them for destruction. T cells recognize infected cells by their altered surface markers, triggering an immune response.
- Tissue Development: Different cell types in an organ express specific surface markers, guiding their spatial organization and function. For example, markers on brain cells guide their connections to form intricate neural networks.
- Cancer Diagnosis: Tumor cells often display unique markers not found in healthy cells. These markers serve as diagnostic tools and potential targets for cancer therapy.
2. Cell-Cell Interactions and Signaling
Surface markers are like handshake points for cellular communication. They act as docking stations for proteins, triggering a cascade of signals within and between cells.
- Cell Adhesion: Specific surface markers on one cell bind to complementary ones on another, facilitating adhesion and forming intricate cell-cell structures like tissues and organs.
- Signal Transduction: Ligand-receptor binding between surface markers on different cells can activate signaling pathways, leading to diverse cellular responses like growth, differentiation, or migration.
- Immune Regulation: T cells recognize specific surface markers on antigen-presenting cells, stimulating the immune response, while other markers on immune cells can dampen inflammation.
3. Cell Adhesion and Migration
Surface markers act like guideposts and anchors for cell movement.
- Directional Migration: Specific surface markers on cells interact with molecules in the extracellular matrix, directing their migration towards specific destinations during development or wound healing.
- Cell Adhesion and Detachment: Dynamic changes in surface marker expression can regulate cell adhesion strength, allowing cells to detach and migrate freely during processes like embryonic development or immune cell trafficking.
- Extracellular Matrix Interaction: Surface markers on cells bind to components of the extracellular matrix, providing structural support and influencing cell behavior.
4. Differential Regulation of Cellular Functions
Surface markers act like switches, controlling specific functions within individual cells.
- Cell Activation and Differentiation: Specific surface markers on stem cells can be targeted by signaling molecules, triggering their differentiation into specialized cell types.
- Enzyme Regulation: Certain surface markers directly regulate the activity of enzymes within cells, influencing metabolic pathways and cellular functions.
- Drug Targeting: Surface markers on diseased cells can be targeted by therapeutic drugs, specifically inhibiting their growth or function.
Flow Cytometry for Surface Marker Detection
In the grand stage of cell biology, where molecules play intricate roles, flow cytometry shines as the spotlight for studying surface markers. It’s a powerful technique that allows scientists to analyze and sort cells based on their unique patterns of protein expression.
The Principle
Flow cytometry works like a sophisticated sorting machine. Cells suspended in a fluid stream pass through a laser beam one by one. Specific antibodies, each conjugated to a unique fluorescent dye, bind to the target surface markers on the cell. As the cell passes through the laser, the dye gets excited and emits a characteristic fluorescent signal. This signal is then detected by sensors, revealing the presence and quantity of the targeted surface marker.
Multiparametric Immunophenotyping
Flow cytometry’s true power lies in its ability to analyze multiple markers simultaneously. By using a cocktail of antibodies, researchers can create a detailed profile of a cell’s surface marker expression, like a multi-colored fingerprint. This technique, called multiparametric immunophenotyping, allows scientists to:
- Distinguish different cell types: Unique combinations of surface markers define cell identities. For example, identifying the presence of CD3 and CD8 on a cell tells us it’s a cytotoxic T lymphocyte.
- Track cell differentiation: As cells mature, their surface marker profile changes. Flow cytometry can track these changes, providing insights into the developmental journey of a cell.
- Study cellular function: Certain surface markers are associated with specific cellular functions. Flow cytometry helps correlate marker expression with cellular activity.
Fluorescence-Activated Cell Sorting (FACS)
Flow cytometry isn’t just about observation; it can also be used to isolate specific cell populations. By combining the analysis with cell sorting capabilities, FACS allows researchers to:
- Purify rare cell populations: For studying specific cell types like HSCs, FACS can isolate them from a complex mixture of blood cells.
- Separate cells based on functional characteristics: Surface markers associated with specific cellular functions can be used to isolate active or resting cells, for example.
Antibodies: The Key Players
The accuracy and specificity of flow cytometry rely heavily on high-quality antibodies. These molecules are designed to bind to specific surface markers with high affinity. By conjugating them to different fluorescent dyes, scientists can create a diverse toolbox for targeted marker detection.
Beyond Flow Cytometry
While flow cytometry reigns supreme, other techniques offer complementary insights:
- Immunohistology: This technique uses antibodies to visualize the localization of surface markers on cells within tissue sections.
- Fluorescence microscopy: This technique allows researchers to observe the spatial distribution of surface markers on individual cells with high resolution.
Clinical Applications of Surface Markers
The development of more fluorochromes and discovery of cell surface markers allow HSPC to be identified prior to pre-transplant in a research setting and later the engraftment of target cell populations are possible to be analyzed in a single test. Also, hematopoietic cells either at its quiescent or activated state can be identified and the aberrancies of blood can be diagnosed. Let’s explore the crucial role they play in clinical settings.
Diagnosis and Classification
- Leukemias: Different leukemia types express distinct surface markers, allowing classification of the disease and proper treatment accordingly. For example, B-cell acute lymphoblastic leukemia (ALL) expresses CD19 and CD38, while T-cell ALL lacks these markers and expresses CD7.
- Lymphomas: Surface markers like CD20 on B cells and CD3 on T cells help distinguish different lymphoma subtypes, guiding treatment strategies.
- Anemias: Aplastic anemia exhibits decreased expression of human leukocyte antigen (HLA) markers on bone marrow cells.
Monitoring Disease Progression and Treatment Response
- Monitoring minimal residual disease (MRD) in leukemia patients after therapy using specific surface marker detection can predict relapse risk and guide treatment adjustments.
- Measuring surface marker expression on immune cells can predict response to immunotherapy in cancer patients.
Identification and Isolation of Specific Cell Populations
- Hematopoietic stem cell transplantation for leukemia and other blood cell disorders relies on identifying and isolating HSCs based on specific surface markers like CD34 and CD133.
- CAR T-cell therapy for cancer involves engineering T cells with receptors targeting specific surface markers on cancer cells.
Development of Targeted Therapies
- Monoclonal antibodies targeting specific surface markers on cancer cells can deliver targeted drugs or toxins directly to the tumor.
- Small-molecule drugs can be designed to interfere with signaling pathways activated by specific surface markers.
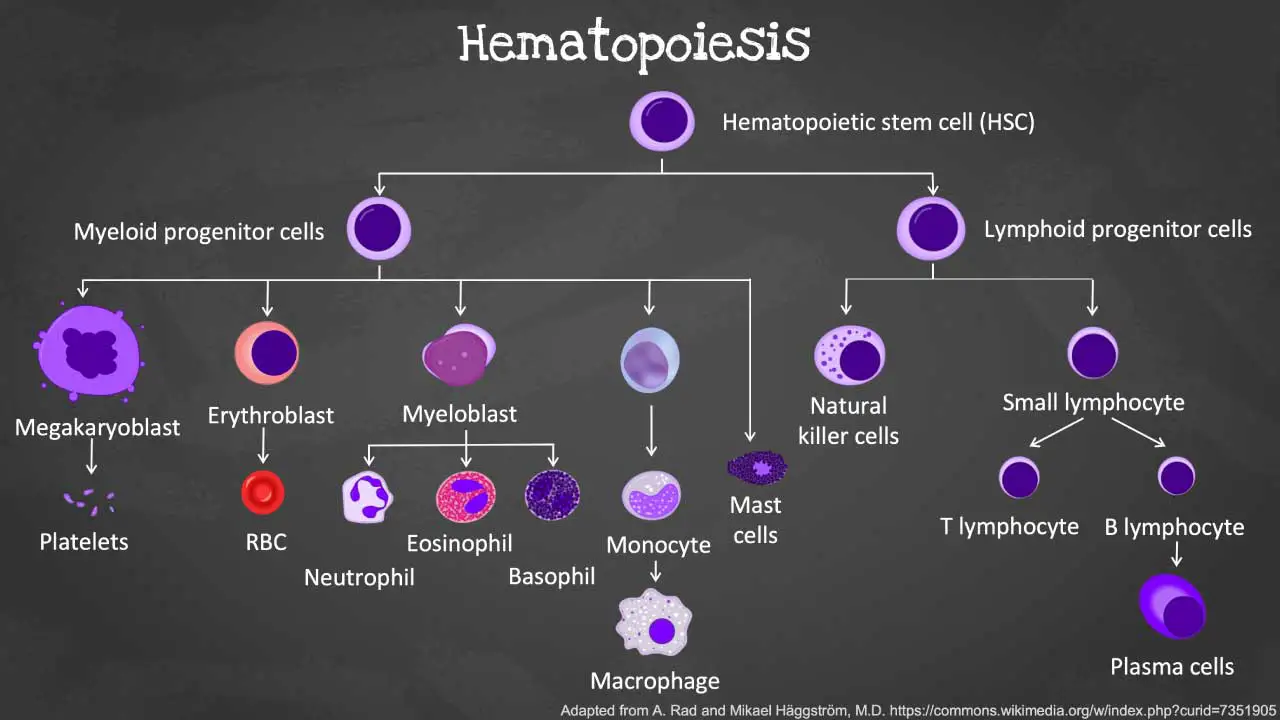
Introduction to Hematopoiesis
Hematopoiesis, pronounced “hee-ma-toh-poy-ee-sis,” is the remarkable process by which your body creates all the different types of blood cells including the red blood cells, white blood cells and platelets. . It’s like having a mini-factory inside your bones, constantly churning out these tiny workers to keep your body functioning smoothly.
During this process of blood production, the different blood cells are derived from the hematopoietic stem and progenitor cells (HSPC) through a tightly coordinated sequence of events. The mother cell from which all blood cells originate from is the hematopoietic stem cell (HSC). Hematopoietic stem cells are multipotent stem cells. Multipotent stem cells are stem cells that can give rise to a limited number of cell types within a specific lineage. They are more differentiated than pluripotent stem cells, which can give rise to all cell types in the body, but less differentiated than unipotent stem cells, which can only give rise to one type of cell. Hematopoietic stem cells (HSCs) have the ability to self-renew, which means they can divide to produce more stem cells and they can differentiate into a limited number of cell types within a specific lineage.
From the HSCs, hematopoietic progenitor cells (HPC) such as the common lymphoid progenitors (CLP), granulocyte-macrophage progenitors (GMP) and megakaryocyte-erythroid progenitors (MEP) arises to give differentiate into hematopoietic precursors, then become increasingly lineage-specific and ultimately differentiate into all lineages of mature hematopoietic cells.
The CLP will differentiate further to develop into T-, B- and natural killer (NK-) cells. These cells are immune cells that are a well-coordinated army defending our body against invaders. The GMPs form granulocyte-macrophage lineage cells such as neutrophils, basophils, eosinophils, mast cells, dendritic cells or monocytes; which are frontline defenders against infections, whereas the megakaryocytes and platelets as well as red blood cells derive from MEPs. Differentially expressed surface markers serve as vital tools for isolating and characterizing HSCs and their lineage-restricted progenitors within the complex hematopoietic hierarchy.
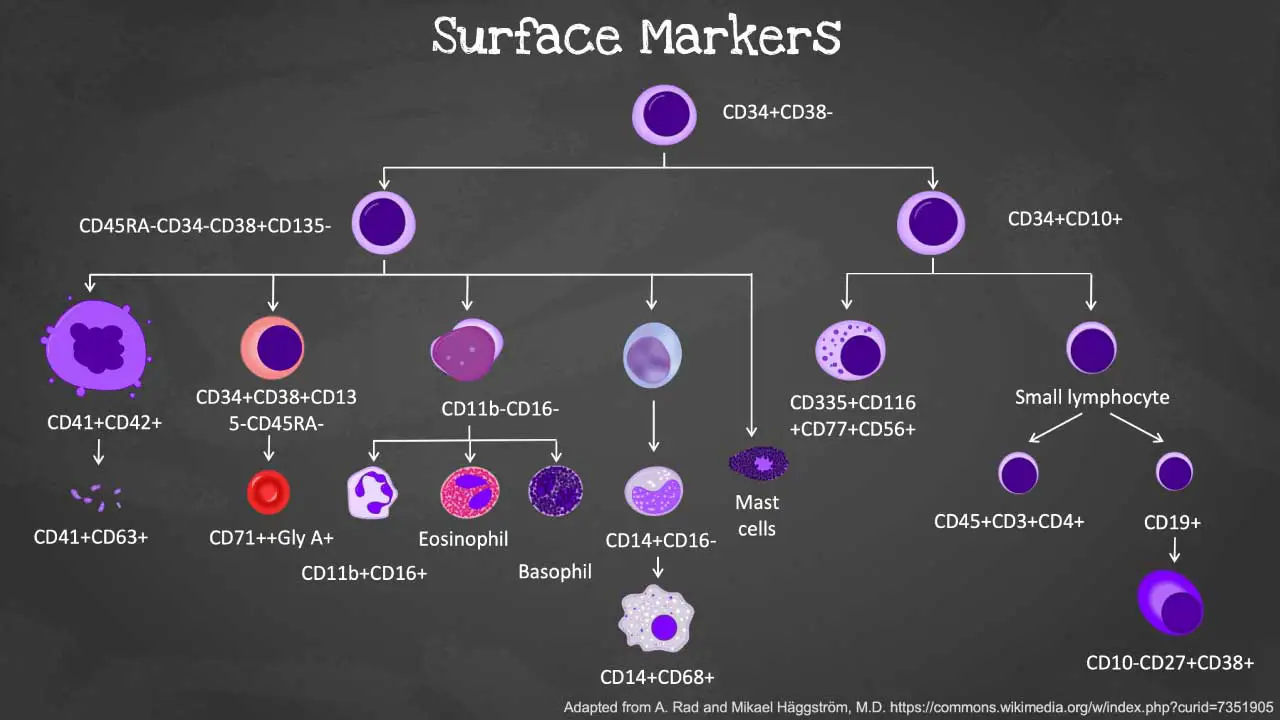
Differential Surface Markers of Hematopoietic or Blood Cells
| Cell type | Surface Markers | References |
| Hematopoietic stem cells | CD34+CD38- -Lin−CD34+CD38−CD90+CD45RA− Lin−CD34+CD38−CD49f+CD45RA−CD90+/− CD34+CD38-CD44+GPI-80+CD90+CD133+CD135+CD3-CD10-CD14-CD19-CD45RA-CD56-CD66b-CD335- | Classical Majeti et al., 2007; Wang, 2012 Qiao et al., 2014 Weeda et al., 2022 |
| Common lymphoid progenitors | CD34+CD10+ Lin−CD34+CD10+ (bone marrow) CD34+CD38-CD45RA+CD7+ (cord blood) | Classical Hokland et al., 1987 Hao et al., 2001 |
| Common myeloid progenitors | CD45RA-CD34+CD19-IL-3R⍺low/-CD110- Lin−CD34+CD38+CD135+CD45RA−CD7−CD10− CD45RA-CD34-CD38+CD135- (BM and cord blood) CD34+CD38+CD123intCD135+CD133lowCD3-CD10-CD11c-CD14-CD19-CD45RA-CD49f-CD56-CD66b-CD90-GPI-80- | Edvardsson et al., 2006 Qiao et al., 2014 Wang, 2012 Weeda et al., 2022 |
| Granulocyte-macrophage progenitors | CD34+CD123+ CD19-CD34+IL-3R⍺low/-CD45RA+CD110- CD34+CD38+CD33+CD45RA+CD117+CD135+HLA-DR+CD3-CD10-CD11c-CD14-CD19-CD56-CD66b-CD90-GPI-80-CD115-CD116-CD123- | Huang et al., 1999 Edvardsson et al., 2006 Weeda et al., 2022 |
| Megakaryocyte-erythroid progenitors | CD34+CD123- CD19-CD34+IL-3Ralow/-CD45RA-CD110+ Lin−CD34+CD38+CD135−CD45RA−CD7−CD10− CD34+CD38+CD135-CD45RA- CD34+CD38+CD123intCD133lowCD3-CD10-CD11c-CD14-CD19-CD45RA-CD56-CD66b-CD90-CD135-GPI-80- | Huang et al., 1999 Edvardsson et al., 2006 Qiao et al., 2014 Wang, 2012 Weeda et al., 2022 |
| Monocyte/dendritic progenitor | CD34+CD38+CD33+CD45RA+CD115+CD117+CD123hiCD135+HLA-DR+CD3-CD10-CD11c-CD14-CD19-CD56-CD66b-CD90-CD116-CD123-GPI-80- | Weeda et al., 2022 |
| Common dendritic progenitor | CD34+CD38+CD11c+CD33+CD45RA+CD116+CD117+CD123hiCD135+HLA-DR+CD3-CD10-CD11c-CD14-CD19-CD56-CD66b-CD90-CD115-GPI-80- | Weeda et al., 2022 |
| T cells | CD45+CD3+CD4+ CD4+ (T helper cells) CD25+CD127+FoxP3+ (regulatory T cells) | O’Gorman and Gelman, 1997 Romagnani, 1991 Fontenot et al., 2003; Yu et al., 2012 |
| Natural killer cells | CD16+ (peripheral blood) CD56+ (mucosal) CD56+ or CD16−CD56− (lymph nodes) | Mandelboim et al., 1999 Reeves et al., 2010 Reeves et al., 2010 |
| B cells | CD19+ (classical) CD10+CD27−CD38+ (immature) CD10−CD27−CD38− (naïve) CD10−CD27+CD38− (memory) CD10−CD27+CD38+ (plasma) | Carter et al., 2002; Wang et al., 2012 Baez et al., 2014; Caraux et al., 2010; Pritz et al., 2015 |
| Monocytes | CD14+ (general) CD14+CD16− (classical) CD14++CD16+ (intermediate) CD14+CD16++ (non-classical) | Martin et al., 1994; Zamani et al., 2013 Passlick et al., 1989; Sulicka et al., 2013 |
| Dendritic cells | MHC II+Lin- CD1c+CD141+ (myeloid derived) CD123+CD303+ CD304+ (plasmacytoid-derived) | Villadangos et al., 2001 Jongbloed et al., 2010 |
| Granulocytes | CD45+CD11b+CD16+ (immature) CD11b─CD16─ (promyelocyte) CD11b+CD16─ (metamyelocyte) CD11b+CD16+ (mature neutrophils) CD66b+ (eosinophil) CD13+CD107a+CD164+ (activated basophils) | Fujimoto et al., 2000; Guerin et al., 2014; Mazzone and Ricevuti, 1995 Fujimoto et al., 2000 Daley et al., 2008 Yoon et al., 2007 Hennersdorf et al., 2005 |
| Erythroid cells | CD71++gly A+ | Rogers et al., 1996 |
| Platelets | PAC-1+CD63+ CD41+CD62P+ CD42b | van Velzen et al., 2012 |
Differential Surface Markers of Leukemias
Adapted from Li W. Flow Cytometry in the Diagnosis of Leukemias. In: Li W, editor. Leukemia [Internet]. Brisbane (AU): Exon Publications; 2022
| Disorders | Surface marker characteristics |
| Precursor B-ALL | CD45dim-negativeCD19+CD10+CD34+CD22+CD38+cytoCD79a+TdT+CD9+CD58+Cd33+CD20-kappa-lambda- |
| Philadelphia-like B-ALL | CRLF2dim-intCD45dim-negativeCD19+CD10+CD34dim-int |
| T-ALL | CD45+CD7+CD4+CD8+CD1a+CD5+Cyto CD3+TdT+CD2+sCD3-CD16-CD56-CD34-αβ-TCR-γδ-TCR- |
| Early-T-precursor (ETP) ALL | CD7+CD34+CD5dimpartialCD45dim-negativeCD4-CD8-CD1a- |
| AML-M1 | CD45dimCD34dimCD117+HLA-DR+CD38+CD13+CD33+CD15+CD123+MPO+CD71-CD64-CD14-lyso- |
| AML-M2 | MPO+CD11b+CD13+CD15+CD33+CD65+HLA-DR+CD34+CD38+CD117+ |
| AML-M3 | CD45intCD117+CD38+CD13+CD33+CD15dimCD64dimpartialCD71partialMPO+lysopartialCD34-HLA-DR-CD114-CD11b-CD11c- |
| AML-M5 | CD45+HLA-DR+CD38+CD33+CD123+CD64+CD11b+CD11c+lyso+CD117-CD34-MPO- |
| AML-M6 | CD71+glycoA+CD36+CD117+CD34-HLA-DR- |
| AML-M7 | CD41+CD61+CD42b+CD34-HLA-DR- |
| AML-M7 with RAM phenotype | CD34+CD56+CD117+CD61dimCD38dim-negativeCD45dim-negativeHLA-DR- |
| Blastic plasmacytoid dendritic cell neoplasm | CD4+CD56+CD23+CD303+CD304+TCF4+TCL1+CD45dim-intCD7+CD33+CD3-CD14-CD19-CD34-lyso-MPO- |
| Burkitt leukemia | CD45+CD19+CD10+CD20+CD38+kapparestriction |
| CLL | CD20+CD22+CD23+FMC-7-sIg+(weak with light chain restriction)CD200+LEF1+ |
| Mantle cell lymphoma | CD20+sIg+CD23-or dimFMC-7+CD200-LEF1- |
| Hairy cell leukemia | CD20+CD11c+CD25+CD103+CD22+sIg+ with light chain restriction FMC-7+CD23-CD5-CD10- |
| Marginal zone lymphoma | Lack of combine positivity for CD11c, CD25, CD103Lack of bright staining for CD20, CD22 |
| Plasma cell leukemia | CD38+CD138+CD19-CD20- |
Conclusion
In conclusion, surface markers are invaluable tools for understanding and regulating hematopoiesis. As research delves deeper and discovers new markers, they pave the way for personalized medicine and developing innovative therapies for blood-related diseases. They play an essential role as different blood cell types express unique surface marker profiles, allowing researchers to distinguish between HSCs, progenitors, and mature cells. This helps map the intricate developmental pathways within the hematopoietic system. Specific markers mediate essential functions like cell adhesion, migration, and signaling, providing insights into the mechanisms governing blood cell development and function. Abnormal expression of certain markers can be used to diagnose and classify blood disorders like leukemias and lymphomas, guiding treatment strategies.
In development of research and innovative therapies, specific markers can be targeted to isolate and manipulate specific cell populations, such as HSCs for transplantation or CAR T-cell therapy, drugs can be designed to target specific markers on diseased cells, offering targeted therapies with fewer side effects and introducing genes encoding specific markers can modify the behavior of blood cells, potentially leading to new treatment options.
Frequently Asked Questions (FAQs)
What are surface markers on a blood test?
When you hear “surface markers on a blood test,” it’s not referring to markers literally present on the surface of your blood itself, but rather on the various cells found within your blood. Specifically, these markers are proteins, carbohydrates, or lipids located on the outer membranes of white blood cells, which play a crucial role in your immune system.
These unique markers, also known as clusters of differentiation (CDs), act like identifying badges for different types of white blood cells. By analyzing the specific combination of markers present on these cells in a blood test, doctors can gain valuable insights into your immune system function and diagnose potential health issues.
Here’s a breakdown of how surface markers help in blood tests:
1. Identifying Cell Types: Different white blood cells (lymphocytes, monocytes, neutrophils, etc.) express distinct sets of surface markers. This allows doctors to count and characterize the different populations of white blood cells in your blood sample, revealing potential imbalances or abnormalities.
2. Diagnosing Disorders: Abnormal expression of certain surface markers can be indicative of various conditions, including:
- Infections: Specific marker patterns may suggest bacterial, viral, or parasitic infections.
- Immune system disorders: Autoimmune diseases and immunodeficiencies often show altered surface marker profiles on different immune cell types.
- Blood cancers: Leukemias and lymphomas have characteristic marker expression patterns that aid in their diagnosis and classification.
3. Monitoring Treatment Response: By tracking changes in surface marker expression over time, doctors can evaluate the effectiveness of treatment for certain conditions like leukemia or monitor immune cell function after organ transplantation.
However, it’s important to understand that:
- Surface marker analysis is often combined with other blood tests and examinations for a comprehensive diagnosis.
- Different types of blood tests use various techniques to assess surface markers, with flow cytometry being a common method.
- Interpreting surface marker results requires expertise and context provided by healthcare professionals.
Are surface markers and CD the same?
No, surface markers and CDs are not exactly the same, but they are closely related:
Surface markers
- These are the broader term, referring to any molecule (protein, carbohydrate, lipid) located on the outer surface of a cell. They can have various functions, like cell-cell communication, adhesion, and immune recognition.
- There are thousands of different surface markers on various cell types throughout the body.
CD (cluster of differentiation)
- This is a nomenclature system used specifically for surface markers on leukocytes (white blood cells).
- Each CD marker has a unique number assigned to it (e.g., CD4, CD8, CD19).
- This system helps standardize the identification and characterization of leukocytes based on their surface marker expression.
The relationship
- Not all surface markers on leukocytes are CDs. Some have different names or haven’t been assigned a CD number yet.
- All CDs, however, are considered surface markers by definition.
What do CD markers mean?
CD markers, short for cluster of differentiation, are a system used to classify and identify specific molecules found on the surface of leukocytes, also known as white blood cells. Each CD marker is assigned a unique number, such as CD4 or CD8, allowing researchers and healthcare professionals to easily refer to them and understand their functions.
Here’s a breakdown of what CD markers mean:
1. Identification and Characterization
Different white blood cell types, like B cells, T cells, and monocytes, express distinct combinations of CD markers. This unique “fingerprint” allows scientists to distinguish different cell types and understand their roles in the immune system.
2. Function and Significance
Each CD marker has a specific function, such as mediating cell-cell communication, adhesion, or activation. Understanding these functions sheds light on how white blood cells work and interact with each other.
3. Diagnosis and Classification
Abnormal expression of specific CD markers can be indicative of various health conditions, including:
- Infections: Certain marker patterns might suggest bacterial, viral, or parasitic infections.
- Immune system disorders: Autoimmune diseases and immunodeficiencies often show altered CD marker profiles on different immune cell types.
- Blood cancers: Leukemias and lymphomas have characteristic marker expression patterns that aid in their diagnosis and classification.
4. Monitoring and Prognosis
By tracking changes in CD marker expression over time, doctors can evaluate the effectiveness of treatment for certain conditions like leukemia or monitor immune cell function after organ transplantation.
5. Further Research and Applications
Researchers are constantly discovering new CD markers and their functions, contributing to our understanding of the immune system and developing novel diagnostic and therapeutic tools.
Can specific surface marker profiles on individual HSCs predict their differentiation potential and therapeutic value in transplantation?
The question of whether specific surface marker profiles on individual HSCs can predict their differentiation potential and therapeutic value in transplantation is a fascinating and actively researched area in hematology. While a definitive answer hasn’t been reached yet, promising developments and ongoing research shed light on the potential.
Current Understanding
- Heterogeneity within HSCs: HSCs, despite appearing alike under a microscope, exhibit significant heterogeneity in their differentiation potential and self-renewal capacity.
- Surface markers capture some variation: Studies have identified various surface markers associated with specific differentiation biases (e.g., towards B-cells, T-cells, or myeloid lineages).
- Limitations exist: Existing marker panels only partially capture the complex differentiation potential of HSCs, and predicting therapeutic value remains challenging.
Promising Directions
- Single-cell analysis: Technologies like single-cell RNA sequencing are revealing deeper insights into individual HSC profiles, including gene expression and functional differences.
- Novel markers: Identifying new surface markers specific to subpopulations of HSCs with distinct properties could improve prediction accuracy.
- Functional assays: Developing assays that test the differentiation potential or therapeutic efficacy of individual HSCs based on their marker profiles would be invaluable.
Challenges and Considerations
- Technical limitations: Single-cell analysis techniques are still evolving, and isolating and analyzing individual HSCs without altering their properties remains a challenge.
- Data interpretation: Integrating vast amounts of data from single-cell analysis with functional assays requires advanced computational and statistical approaches.
- Ethical considerations: Manipulating HSCs based on their predicted properties raises ethical concerns that need careful consideration.
Overall, while predicting individual HSC differentiation and therapeutic value solely based on surface markers remains elusive, continued research in single-cell analysis, novel marker identification, and functional assays holds tremendous promise for personalized medicine in stem cell transplantation.
Additional Insights
- Studying epigenetics and metabolic profiles alongside surface markers might provide a more comprehensive understanding of HSC potential.
- Integrating information from diverse data sources using machine learning algorithms could improve prediction accuracy.
- Collaboration between researchers, clinicians, and ethicists is crucial to ensure responsible development and application of these technologies.
How do microenvironmental cues influence surface marker expression during hematopoiesis and how can this be leveraged for therapeutic targeting?
The journey of a blood cell from a humble hematopoietic stem cell (HSC) to its specific mature form is guided by a complex interplay between intrinsic factors and extrinsic cues within the bone marrow microenvironment (BME). Among these extrinsic cues, microenvironmental signals play a crucial role in regulating the expression of surface markers on HSCs and their progeny, directing their differentiation, proliferation, and survival.
How do microenvironmental cues influence surface marker expression?
Various factors in the BME
- Growth factors: Cytokines like SCF, IL-7, and G-CSF interact with specific receptors on HSCs, influencing their survival, proliferation, and commitment towards specific lineages. This often leads to changes in surface marker expression reflecting their changing potential.
- Adhesion molecules: Interactions between HSCs and stromal cells through molecules like VCAM-1 and N-cadherin provide physical contact and critical signals that impact gene expression and, consequently, surface marker profiles.
- Metabolic factors: Hypoxia (low oxygen) within the BME promotes self-renewal of HSCs, while increased oxygen levels favor differentiation. This metabolic shift can also influence the expression of certain markers.
- Niche signals: Specialized niches within the BME provide unique signals to HSCs depending on their location. For example, osteoblastic and endothelial niches offer distinct cues that influence marker expression and differentiation potential.
Leveraging this knowledge for therapeutic targeting
Understanding how microenvironmental cues influence surface marker expression opens exciting avenues for therapeutic strategies:
- Manipulating BME factors: By introducing specific growth factors or mimicking niche signals, scientists can potentially guide HSCs towards desired differentiation pathways, overcoming limitations in current HSC transplantations.
- Targeting marker-specific pathways: Identifying signaling pathways activated by specific microenvironmental cues that lead to altered marker expression could help develop drugs that modulate these pathways, influencing HSC behavior for therapeutic benefit.
- Designing marker-based therapies: Utilizing specific surface markers expressed on HSCs or their progeny allows for targeted delivery of drugs or gene therapies, enhancing treatment efficacy and reducing side effects.
Challenges and future directions
While promising, this field requires further exploration:
- Complexity of the BME: Deciphering the intricate and dynamic interplay of various factors within the BME remains a challenge.
- Limited understanding of signaling pathways: More research is needed to understand the precise downstream effects of microenvironmental cues on specific surface marker expression and HSC function.
- Translational challenges: Bridging the gap between in vitro studies and clinical applications requires careful evaluation of safety and efficacy in animal models and clinical trials.
Can novel technologies like single-cell proteomics and spatial transcriptomics reveal new and functionally relevant surface markers in leukemia diagnosis and therapy?
Absolutely! Novel technologies like single-cell proteomics and spatial transcriptomics hold immense potential in uncovering new and functionally relevant surface markers for leukemia diagnosis and therapy.
Limitations of Current Approaches
Traditional bulk analysis of leukemic cells often provides an average picture, masking the heterogeneity within the tumor microenvironment. This can lead to inaccurate diagnoses and ineffective therapies.
The Promise of Single-Cell Proteomics
- Unveiling Hidden Heterogeneity: By analyzing protein expression on individual cells, single-cell proteomics can reveal distinct subpopulations within the leukemia and identify rare cell types with specific features, aiding in targeted therapies.
- Going Beyond Genes: Unlike traditional RNA sequencing, single-cell proteomics directly measures protein levels, providing a more accurate picture of cellular activity and function.
- Discovering Novel Markers: This technology can identify previously unknown surface markers associated with specific subpopulations or drug resistance mechanisms, opening doors for personalized diagnosis and targeted therapies.
Spatial Transcriptomics
- Mapping the Tumor Microenvironment: This technology creates a spatial map of gene expression within the tumor tissue, revealing how different cell types interact and influence each other.
- Identifying Niche-Specific Markers: By understanding the location of specific cell types and their communication, spatial transcriptomics can help identify surface markers specific to their microenvironment, providing insights into drug resistance and potential therapeutic targets.
- Building a Comprehensive Picture: Integrating data from single-cell proteomics with spatial transcriptomics can paint a holistic picture of the tumor microenvironment, leading to more effective treatment strategies.
Challenges and Future Directions
- Data Analysis and Integration: Analyzing and interpreting massive datasets from these technologies requires advanced computational tools and expertise.
- Clinical Translation: Bridging the gap between basic research and clinical applications necessitates further validation and development of robust assays.
- Ethical Considerations: The potential for personalized medicine based on these technologies raises complex ethical questions that need careful discussion.
Overall, single-cell proteomics and spatial transcriptomics are revolutionizing our understanding of leukemia and its complexity. By revealing new and functionally relevant surface markers, these technologies hold immense promise for improving diagnosis, developing targeted therapies, and ultimately conquering this challenging disease.
Additionally, other advancements like CRISPR-Cas9 genome editing have potential to create designer immune cells targeting specific surface markers on leukemic cells. This opens doors for novel immunotherapies with increased efficacy and reduced side effects.
How can we overcome the limitations of current antibodies targeting surface markers for effective and specific drug delivery in leukemias?
Despite the promise of using surface markers on leukemic cells for targeted drug delivery, current antibody-based approaches face several limitations:
1. Heterogeneity: Leukemia is a diverse disease with various subtypes and within-patient heterogeneity. Targeting a single surface marker might not capture all relevant cell populations.
2. Off-target effects: Antibodies can bind to unintended healthy cells expressing similar markers, leading to toxicity and side effects.
3. Internalization and drug release: Even with specific binding, efficient drug delivery requires antibody internalization, proper drug release within the cell, and overcoming drug resistance mechanisms.
4. Immunogenicity: Repeated antibody administration can trigger immune responses, reducing their effectiveness and posing safety concerns.
5. Cost and complexity: Designing, manufacturing, and delivering antibody-based therapies can be expensive and complex.
Here are some promising strategies to overcome these limitations:
1. Combination therapy: Targeting multiple surface markers with antibody cocktails or bispecific antibodies can improve specificity and capture heterogeneous populations.
2. Antibody engineering: Modifying antibodies with polymers, toxins, or drugs can enhance their potency, improve internalization, and overcome drug resistance.
3. Nanoparticle-based delivery: Utilizing nanoparticles as delivery vehicles can encapsulate various drugs, target specific markers, and offer controlled release mechanisms.
4. CAR T-cell therapy: Engineering T cells with chimeric antigen receptors (CARs) recognizing specific leukemia markers can induce potent and targeted cell killing.
5. Small molecule inhibitors: Developing small molecules that target signaling pathways downstream of surface markers can offer broad-spectrum targeting and avoid antibody-associated limitations.
6. Artificial intelligence and machine learning: Utilizing these tools to analyze patient data and predict optimal targets based on individual tumor profiles can personalize treatment strategies.
7. Overcoming immunogenicity: Strategies like humanization of antibodies, immunosuppressive drugs, and immune checkpoint inhibitors can reduce antibody-related immune responses.
8. Exploring alternative targeting approaches: Utilizing aptamers, peptides, or affibodies as targeting moieties can offer advantages over traditional antibodies in terms of cost, flexibility, and immunogenicity.
By exploring these innovative approaches and overcoming the limitations of current strategies, we can move towards more effective and specific drug delivery in leukemias, ultimately improving patient outcomes and survival rates.
Can surface markers on immune cells be manipulated to enhance their anti-tumor activity in CAR T-cell therapy for leukemias?
Absolutely! Manipulating surface markers on immune cells, particularly in the context of CAR T-cell therapy, holds immense potential for enhancing their anti-tumor activity in leukemias. Here’s how:
Current Challenges
- Limited tumor recognition: CAR T-cells are often designed to target a single marker, which can lead to tumor escape due to antigen loss or heterogeneity.
- T cell exhaustion: The harsh tumor microenvironment can exhaust CAR T-cells, reducing their efficacy and persistence.
- On-target, off-tumor toxicity: Targeting certain markers might also affect healthy tissues expressing the same marker, leading to side effects.
Strategies for Manipulation
- Multi-targeted CAR T-cells: Engineering CAR T-cells to recognize multiple tumor-specific markers simultaneously can address heterogeneity and prevent antigen escape.
- Co-stimulatory molecules: Introducing additional signaling molecules alongside the CAR can enhance T-cell activation and persistence, overcoming exhaustion.
- Suicide genes: Incorporating genes that can selectively eliminate CAR T-cells if needed minimizes risks associated with on-target, off-tumor toxicity.
- Checkpoint inhibitors: Combining CAR T-cell therapy with drugs that block inhibitory signals in the tumor microenvironment can reignite T-cell activity and improve efficacy.
- Arming CAR T-cells: Modifying CAR T-cells to secrete cytokines or cytotoxic molecules can directly increase their tumor-killing potential.
Emerging Technologies
- CRISPR-Cas9: Precisely editing genes within T-cells allows for targeted manipulation of surface markers or signaling pathways to enhance their anti-tumor properties.
- Nanoparticle-based delivery: Delivering therapeutic molecules or genetic instructions specifically to CAR T-cells can further improve their functionality and efficacy.
Considerations and Future Directions
- Safety and efficacy: Balancing optimal tumor targeting with minimizing potential side effects remains a critical consideration.
- Clinical translation: Translating pre-clinical success into effective and personalized therapies for patients requires meticulous clinical trials and evaluation.
- Ethical considerations: Careful discussion and ethical frameworks are necessary as personalized medicine based on surface marker manipulation raises complex questions.
Can circulating tumor cells (CTCs) isolated based on their surface marker profiles provide valuable prognostic and therapeutic information in leukemia patients?
The potential of circulating tumor cells (CTCs) isolated based on their surface marker profiles for providing valuable prognostic and therapeutic information in leukemia patients is an exciting and actively researched area. Here’s a breakdown of the current understanding and future possibilities.
Potential Advantages
- Liquid biopsy: CTCs offer a “liquid biopsy” alternative to invasive bone marrow biopsies, providing minimally invasive access to tumor information.
- Heterogeneity: Analyzing individual CTCs with specific surface marker profiles can capture the heterogeneity within a leukemia and identify subpopulations with distinct behavior and therapeutic vulnerabilities.
- Minimal residual disease (MRD) detection: Sensitive detection of rare CTCs even after treatment completion (MRD) can predict relapse risk and guide treatment adjustments.
- Therapeutic targeting: Identifying unique surface markers on CTCs could pave the way for targeted therapies or personalized medicine approaches.
Current Challenges
- Rarity and heterogeneity: CTCs are extremely rare in blood, and leukemia exhibits significant heterogeneity, making isolation and analysis challenging.
- Limited sensitivity and specificity: Existing technologies might not be able to reliably distinguish leukemic CTCs from healthy cells or other blood-borne tumor cells.
- Functional significance: Understanding the functional significance of specific surface marker profiles on CTCs in the context of leukemic progression and relapse is still evolving.
- Clinical validation: More research and clinical trials are needed to fully validate the prognostic and therapeutic value of CTC analysis in leukemia.
Promising Directions
- Advanced technologies: Single-cell analysis, microfluidic devices, and machine learning algorithms are being developed to improve sensitivity, specificity, and capture of relevant information from CTCs.
- Combination of markers: Utilizing panels of surface markers specific to different leukemia subtypes or disease stages could enhance diagnostic and prognostic accuracy.
- Functional assays: Integrating CTC analysis with functional assays to assess drug sensitivity or resistance could guide personalized treatment strategies.
- Early detection and MRD monitoring: Utilizing CTC analysis for early detection of minimal residual disease could improve treatment outcomes and prevent relapse.
How can we integrate information from surface markers with other omics data (e.g., genomics, metabolomics) to develop more comprehensive models of leukemia progression and treatment response?
Integrating information from surface markers with other omics data is a powerful approach to gain a more comprehensive understanding of leukemia progression and treatment response. By combining diverse data sets, researchers can create more accurate and informative models that could eventually lead to personalized medicine strategies. Here are some promising avenues for such integration.
Benefits of integration
- Unveiling complex interactions: Omics data provides insights into different layers of cellular information, from genes and proteins to metabolites. Integrating surface marker data with these can uncover how genetic mutations, metabolic pathways, and protein expression interact, influencing disease progression and drug response.
- Identifying novel targets: By analyzing correlations between surface markers and other omics data, researchers can identify potential new therapeutic targets that were not evident from looking at each data set individually.
- Personalized medicine: Integrating information from individual patients’ surface markers and other omics data can pave the way for personalized treatment plans tailored to their specific disease characteristics and predicted response to specific drugs.
- Improved risk stratification: Integrating omics data can help better categorize patients into risk groups based on their molecular profiles, allowing for more targeted prevention and treatment strategies.
Challenges and considerations
- Data complexity: Analyzing and integrating massive datasets from multiple omics platforms requires advanced computational tools and expertise.
- Data standardization: Omics data generated from different platforms and labs can vary in format and quality, necessitating standardization for effective integration.
- Biological interpretation: Extracting meaningful biological insights from complex multi-omics data requires robust statistical methods and domain knowledge.
- Ethical considerations: Integrating patient-specific omics data raises ethical concerns regarding privacy, data ownership, and potential discrimination based on genetic information.
Promising approaches
- Machine learning and AI: These tools can help analyze vast omics datasets, identify hidden patterns, and build predictive models of disease progression and drug response.
- Network biology: Constructing networks that integrate different omics data can reveal how various molecules and pathways interact, leading to a deeper understanding of disease mechanisms.
- Single-cell analysis: Analyzing individual cells with multi-omics approaches can capture heterogeneity within tumors and identify rare cell populations with specific properties.
Overall, integrating surface marker data with other omics information holds immense potential for developing more comprehensive models of leukemia and guiding personalized treatment strategies. Addressing the challenges and ethical considerations while utilizing advanced analytical tools and collaborative research efforts will be crucial to unlocking the full potential of this promising approach.
In addition to the points mentioned above, it’s important to consider the specific type of leukemia being investigated, as different subtypes might have unique molecular signatures and require tailored multi-omics approaches. By continuously refining methodologies and exploring novel tools, the integration of surface marker and other omics data promises to revolutionize our understanding and treatment of leukemia in the future.
Disclaimer: This article is intended for informational purposes only and is specifically targeted towards medical students. It is not intended to be a substitute for informed professional medical advice, diagnosis, or treatment. While the information presented here is derived from credible medical sources and is believed to be accurate and up-to-date, it is not guaranteed to be complete or error-free. See additional information.
References
- Majeti R, Park CY, Weissman IL. Identification of a hierarchy of multipotent hematopoietic progenitors in human cord blood. Cell Stem Cell. 2007;1(6):635-45.
- Qiao W, Wang W, Laurenti E, Turinsky AL, Wodak SJ, Bader GD, et al. Intercellular network structure and regulatory motifs in the human hematopoietic system. Mol Syst Biol. 2014;10:741.
- Wang X, Shok, J., Edinger, M., Warner, N., Bush-Donovan, C. Multiparametric Immunophenotyping of Human Hematopoietic Stem Cells and Progenitors Cells by Flow Cytometry. BD Biosciences Application Note. 2012:1-16.
- Weeda, V.; Mestrum, S.G.C.; Leers, M.P.G. Flow Cytometric Identification of Hematopoietic and Leukemic Blast Cells for Tailored Clinical Follow-Up of Acute Myeloid Leukemia. Int. J. Mol. Sci. 2022, 23, 10529. https://doi.org/10.3390/ijms231810529
- Hokland P, Hokland M, Daley J, Ritz J. Identification and cloning of a prethymic precursor T lymphocyte from a population of common acute lymphoblastic leukemia antigen (CALLA)-positive fetal bone marrow cells. J Exp Med. 1987;165(6):1749-54
- Hao QL, Zhu J, Price MA, Payne KJ, Barsky LW, Crooks GM. Identification of a novel, human multilymphoid progenitor in cord blood. Blood. 2001;97(12):3683-90.
- Edvardsson L, Dykes J, Olofsson T. Isolation and characterization of human myeloid progenitor populations–TpoR as discriminator between common myeloid and megakaryocyte/erythroid progenitors. Exp Hematol. 2006;34(5):599-609.
- Huang S, Chen Z, Yu JF, Young D, Bashey A, Ho AD, et al. Correlation between IL-3 receptor expression and growth potential of human CD34(+) hematopoietic cells from different tissues. Stem cells. 1999;17(5):265-72.
- O’Gorman MR, Gelman R. Inter- and intrainstitutional evaluation of automated volumetric capillary cytometry for the quantitation of CD4- and CD8-positive T lymphocytes in the peripheral blood of persons infected with human immunodeficiency virus. Site Investigators and the NIAID New CD4 Technologies Focus Group. Clin Diagn Lab Immunol. 1997;4(2):173-9.
- Romagnani S. Type 1 T helper and type 2 T helper cells: functions, regulation and role in protection and disease. Int J Clin Lab Res. 1991;21(2):152-8.
- Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4(4):330-6.
- Yu N, Li X, Song W, Li D, Yu D, Zeng X, et al. CD4(+)CD25 (+)CD127 (low/-) T cells: a more specific Treg population in human peripheral blood. Inflammation. 2012;35(6):1773-80.
- Mandelboim O, Malik P, Davis DM, Jo CH, Boyson JE, Strominger JL. Human CD16 as a lysis receptor mediating direct natural killer cell cytotoxicity. Proc Natl Acad Sci U S A. 1999;96(10):5640-4.
- Reeves RK, Gillis J, Wong FE, Yu Y, Connole M, Johnson RP. CD16- natural killer cells: enrichment in mucosal and secondary lymphoid tissues and altered function during chronic SIV infection. Blood. 2010;115(22):4439-46.
- Wang K, Wei G, Liu D. CD19: a biomarker for B cell development, lymphoma diagnosis and therapy. Exp Hematol Oncol. 2012;1(1):36.
- Carter RH, Wang Y, Brooks S. Role of CD19 signal transduction in B cell biology. Immunol Res. 2002;26(1-3):45-54.
- Caraux A, Klein B, Paiva B, Bret C, Schmitz A, Fuhler GM, et al. Circulating human B and plasma cells. Age-associated changes in counts and detailed characterization of circulating normal CD138- and CD138+ plasma cells. Haematologica. 2010;95(6):1016-20.
- Baez A, Alvarez-Laderas I, Piruat JI, Caballero-Velazquez T, Barbado MV, Millan-Ucles A, et al. The CD27 memory B cells display changes in the gene expression pattern in elderly individuals. Immunology. 2014;144(3):399-404.
- Pritz T, Lair J, Ban M, Keller M, Weinberger B, Krismer M, et al. Plasma cell numbers decrease in bone marrow of old patients. Eur J Immunol. 2015;45(3):738-46.
- Zamani F, Zare Shahneh F, Aghebati-Maleki L, Baradaran B. Induction of CD14 Expression and Differentiation to Monocytes or Mature Macrophages in Promyelocytic Cell Lines: New Approach. Adv Pharm Bull. 2013;3(2):329-32.
- Martin TR, Mongovin SM, Tobias PS, Mathison JC, Moriarty AM, Leturcq DJ, et al. The CD14 differentiation antigen mediates the development of endotoxin responsiveness during differentiation of mononuclear phagocytes. J Leukoc Biol. 1994;56(1):1-9.
- Passlick B, Flieger D, Ziegler-Heitbrock HW. Identification and characterization of a novel monocyte subpopulation in human peripheral blood. Blood. 1989;74(7):2527-34.
- Sulicka J, Surdacki A, Mikolajczyk T, Strach M, Gryglewska B, Cwiklinska M, et al. Elevated markers of inflammation and endothelial activation and increased counts of intermediate monocytes in adult survivors of childhood acute lymphoblastic leukemia. Immunobiology. 2013;218(5):810-6.
- Villadangos JA, Cardoso M, Steptoe RJ, van Berkel D, Pooley J, Carbone FR, et al. MHC class II expression is regulated in dendritic cells independently of invariant chain degradation. Immunity. 2001;14(6):739-49.
- Jongbloed SL, Kassianos AJ, McDonald KJ, Clark GJ, Ju X, Angel CE, et al. Human CD141+ (BDCA-3)+ dendritic cells (DCs) represent a unique myeloid DC subset that cross-presents necrotic cell antigens. J Exp Med. 2010;207(6):1247-60.
- Fujimoto H, Sakata T, Hamaguchi Y, Shiga S, Tohyama K, Ichiyama S, et al. Flow cytometric method for enumeration and classification of reactive immature granulocyte populations. Cytometry. 2000;42(6):371-8.
- Guerin E, Orabona M, Raquil MA, Giraudeau B, Bellier R, Gibot S, et al. Circulating immature granulocytes with T-cell killing functions predict sepsis deterioration*. Crit Care Med. 2014;42(9):2007-18.
- Mazzone A, Ricevuti G. Leukocyte CD11/CD18 integrins: biological and clinical relevance. Haematologica. 1995;80(2):161-75.
- Daley JM, Thomay AA, Connolly MD, Reichner JS, Albina JE. Use of Ly6G-specific monoclonal antibody to deplete neutrophils in mice. J Leukoc Biol. 2008;83(1):64-70.
- Yoon J, Terada A, Kita H. CD66b regulates adhesion and activation of human eosinophils. J Immunol. 2007;179(12):8454-62.
- Hennersdorf F, Florian S, Jakob A, Baumgartner K, Sonneck K, Nordheim A, et al. Identification of CD13, CD107a, and CD164 as novel basophil-activation markers and dissection of two response patterns in time kinetics of IgE-dependent upregulation. Cell Res. 2005;15(5):325-35.
- van Velzen JF, Laros-van Gorkom BAP, Pop GAM, van Heerde WL. Multicolor flow cytometry for evaluation of platelet surface antigens and activation markers. Thromb Res. 2012;130(1):92-8.
- Rogers CE, Bradley MS, Palsson BO, Koller MR. Flow cytometric analysis of human bone marrow perfusion cultures: erythroid development and relationship with burst-forming units-erythroid. Exp Hematol. 1996;24(5):597-604.
- Li W. Flow Cytometry in the Diagnosis of Leukemias. In: Li W, editor. Leukemia [Internet]. Brisbane (AU): Exon Publications; 2022 Oct 16. Chapter 4.