Introduction
Beta thalassemia conventional PCR protocol is one of diagnostic laboratory methods used to identify the specific mutation involved in beta-thalassemia. Beta thalassemia is a group of inherited blood disorders characterized by a reduced or absent production of beta-globin, a protein chain that makes up hemoglobin, the oxygen-carrying protein in red blood cells. This deficiency leads to the formation of abnormally small, pale red blood cells (anemia), which can cause a range of symptoms, including fatigue, weakness, pale skin, and shortness of breath.
There are two main types of beta thalassemia: beta thalassemia major and beta thalassemia minor. Beta thalassemia major, also known as Cooley’s anemia, is the most severe form of the disorder. It is caused by mutations in both copies of the beta-globin gene and typically manifests in early childhood. Symptoms of beta thalassemia major can be severe and may include frequent blood transfusions, bone deformities, and heart problems.
Laboratory investigations play a crucial role in diagnosing and monitoring beta thalassemia. These tests can help determine the type and severity of the disorder, assess the effectiveness of treatment, and identify potential complications.
DNA analysis, such as gene sequencing, can identify the specific mutations in the beta-globin gene that cause beta-thalassemia. This information can be used for genetic counselling and prenatal diagnosis.
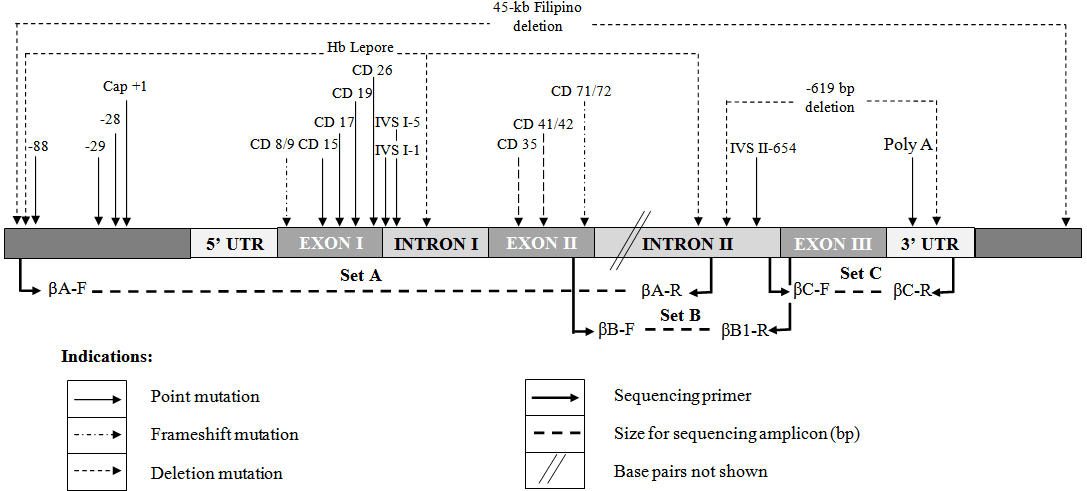
The purpose of this PCR protocol is to amplify the whole β-globin gene as PCR products using conventional PCR protocol so that they can be sent for direct sequencing to identify unknown beta thalassemia mutations. Three segments of amplicons encompassing the entire 1.6 kb of β-globin gene including the 5’ and 3’ of the untranslated region were expected to cover all possible mutations occurring in the β-globin gene.
Principle of Conventional PCR Protocol
Conventional polymerase chain reaction (PCR) is a technique used to amplify small segments of DNA into millions or billions of copies using a conventional thermal cycler . It is a powerful tool in molecular biology and is used in a wide range of applications, including:
- Research: Conventional PCR protocol is used to study DNA from a variety of sources, including viruses, bacteria, plants, and animals. It can be used to identify new genes, study gene expression, and detect mutations.
- Diagnostics: Conventional PCR protocol is used to diagnose a variety of diseases, including infectious diseases, genetic disorders, and cancer.
- Forensic science: Conventional PCR protocol is used to identify individuals from DNA samples found at crime scenes.
Conventional PCR protocol is a highly sensitive and specific technique. It can be used to amplify DNA from even a few cells or from DNA that is degraded or damaged. Conventional PCR protocol is also a relatively fast and easy technique to perform.
Here is a more detailed explanation of the steps involved in conventional PCR protocol:
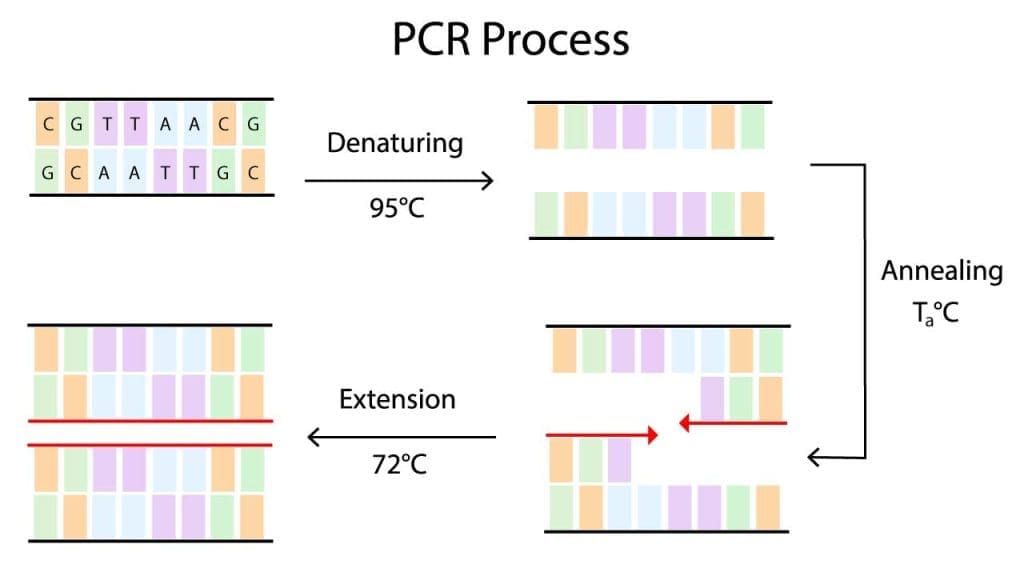
Denaturation
Denaturation is the process of separating the two strands of double-stranded DNA. This is done by heating the DNA to a high temperature, typically 95 degrees Celsius. The high temperature breaks the hydrogen bonds that hold the two strands of DNA together.
Annealing
Annealing is the process of binding the primers to the template DNA. Primers are short oligonucleotide sequences that are complementary to the ends of the target DNA sequence. Primers are important because they help to ensure that the DNA polymerase only amplifies the desired DNA sequence.
Extension
Extension is the process of synthesizing new DNA strands complementary to the template DNA and primers. This is done by adding DNA polymerase to the reaction mixture. DNA polymerase is an enzyme that can synthesize new DNA strands using a template strand and a primer.
The conventional PCR cycle is repeated 20-40 times, resulting in an exponential amplification of the target DNA sequence. At the end of the conventional PCR reaction, the amplified DNA can be analyzed or used in other downstream applications.
Materials
- PCR master mix kit
- Primers
- Pipet tips with aerosol barrier
- Pipettes
- Vortex
- Nuclease-free water
- Genomic DNA
- 1.5 ml microfuge tubes
- 0.2 ml PCR tube/strips
- Thermalcycler
- Gloves
- 70% alcohol (ethanol/isopropanol) spray
Protocol
Table 1. Primer sequences for β-globin gene sequencing.
| Primer ID | Primer sequences (5’ to 3’) | Location: bases (Appendix 3.3c) | Ta(°C) | Amplicon size (bp) |
| HUMHBB U01317: | ||||
| βA – F | CGA TCT TCA ATA TGC TTA CCA A | 61830-61851 | 60 | 916 |
| βA – R | CAT TCG TCT GTT TCC CAT TCT A | 62745-62725 | ||
| βB – F | GCA CGT GGA TCC TGA GAA CT | 62607-62626 | 68 | 902 |
| βB – R | CAC ACA GAC CAG CAC GTT G | 63508-63490 | ||
| βC – F | GCT AAT CAT GTT CAT ACC TCT T | 63445-63466 | 62 | 855 |
| βC – R | CAG ATT CCG GGT CAC TGT G | 64299-64281 |
*The concentration of each primer pair is 4 pmol.
- Wipe down the PCR work bench and pipettes with alcohol spray and turn on UV light for 10 minutes before starting any work.
- Thaw samples and reagents by hand, agitate and spin down. All components should be mixed and spun down prior to pipetting.
- Next, prepare a master mix for the PCR reaction in a 1.5 mL microcentrifuge tube on ice as shown in Table 2.
Table 2. Preparation of Master Mix for PCR reaction.
| Component | Each reaction(50 μL) | Final concentration |
| PCR Master mix* | 25 µL | 1× |
| Primer mixes (Forward and reverse) 10mM stock for each primers | 4 µL | 0.4 µM for each primer |
| Nuclease free water | 18 µL | – |
| Template DNA (can be added later e.g. Step 4) | 3.0 µL | ~300 ng |
| Total | 50 µL |
- Mix master mix gently and spin down using a microcentrifuge.
- Add 47 µL of master mix to each respective PCR tube with pre-loaded 3 µL of template DNA on ice.
- Set the cycling conditions or use the Beta Sequence A/B/C (depending on the set being used) programme in the PCR Thermal Cycler machine as shown in Table 3 and start the run.
Table 3: Thermocycling conditions
| Step | Temperature (°C) | Time (minute(s)) |
| Initial Denaturation | 95°C | 10 minutes |
| Cycling 30 cycles | 95°C | 1 minute |
| Ta °C | 1 minute | |
| 72°C | 1 minute 30 seconds | |
| Final Extension | 72°C | 10 minutes |
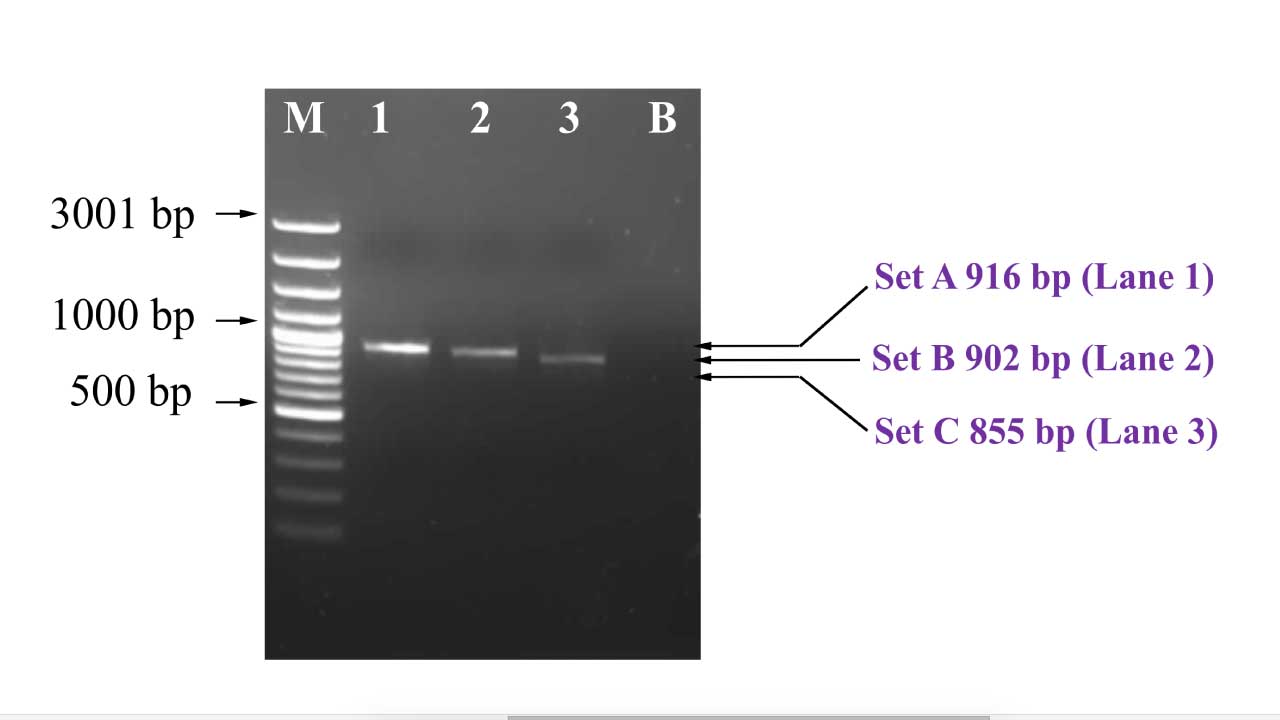
- Proceed to gel electrophoresis.
- The PCR products should be purified before using it for sequencing.
Interpretation
This gel electrophoresis image reveals the distinct bands representing successful amplification of the entire beta globin gene, which will be used for downstream sequencing protocols after PCR clean up protocol.
Troubleshooting
Conventional PCR can be a delicate process, and various factors can influence its success. Here are some common issues and potential solutions:
No or Low Product Yield
- Insufficient template DNA: Increase template amount or use a more sensitive detection method.
- Primer problems: Check primer sequences, concentrations, and annealing temperature. Design new primers if necessary.
- Enzyme issues: Verify enzyme activity and concentration. Try a different polymerase.
- Inhibitors: Purify DNA thoroughly or dilute template.
Non-specific Amplification
- Primer dimer formation: Optimize primer concentrations and annealing temperature. Consider using hot start polymerase.
- Incorrect annealing temperature: Optimize annealing temperature using a gradient PCR. Lower annealing temperature allows for more unspecific binding.
- Contamination: Use proper aseptic techniques and decontamination procedures.
- Incorrect MgCl2 concentration: High MgCl2 concentration will lead to more unspecific binding.
Weak or Faint Bands
- Low template concentration: Increase template amount or number of cycles.
- Inefficient amplification: Optimize PCR conditions (annealing temperature, MgCl2 concentration).
- Primer limitations: Redesign primers or use different primer pairs.
Smear or Multiple Bands
- Template degradation: Use high-quality DNA and avoid excessive freeze-thaw cycles.
- Non-specific amplification: Optimize PCR conditions (annealing temperature, MgCl2 concentration).
- Primer dimer formation: Optimize primer concentrations and annealing temperature.
No Amplification
- PCR mix errors: Double-check all components and their concentrations.
- Thermocycler malfunction: Verify thermocycler performance.
Additional Tips
- Positive and negative controls: Include these in every PCR experiment.
- Optimize reaction components: Experiment with different concentrations of primers, dNTPs, and MgCl2.
- Consider PCR additives: Some additives can improve amplification efficiency.
- Regular maintenance: Clean thermocycler and pipettes regularly.
Remember: Troubleshooting PCR often involves a systematic approach. By carefully analyzing the problem and implementing the appropriate solutions, you can significantly improve your PCR success rate.
Frequently Asked Questions (FAQs)
What are the different types of PCR available?
PCR, or Polymerase Chain Reaction, is a technique used to amplify DNA sequences. There are numerous variations of PCR, each with specific applications. Here are some of the most common types:
Basic PCR Types
- Standard Conventional PCR: The foundational method for amplifying DNA.
- Real-time PCR (qPCR): Quantifies DNA by monitoring amplification in real-time using fluorescent dyes.
- Reverse Transcription PCR (RT-PCR): Converts RNA into DNA (cDNA) before amplification.
Specialized PCR Techniques
- Nested PCR: Increases specificity by using two sets of primers in sequential PCR reactions.
- Multiplex PCR: Amplifies multiple DNA targets simultaneously in a single reaction.
- Long-range PCR: Amplifies longer DNA fragments than standard PCR.
- Digital PCR: Partitions a sample into many small droplets, allowing for absolute quantification of DNA.
- Colony PCR: Screens bacterial colonies for specific genes.
- In situ PCR: Amplifies DNA directly in cells or tissues.
- Hot Start PCR: Improves specificity by preventing primer annealing before the reaction reaches optimal temperature.
Other PCR Variants
- Methylation-specific PCR (MSP): Detects DNA methylation patterns.
- Assembly PCR: Joins multiple DNA fragments together.
- Asymmetric PCR: Produces single-stranded DNA.
- Differential Display PCR: Identifies gene expression differences.
Disclaimer: This protocol is intended for informational purposes only and may need to be modified depending on the specific laboratory procedures and patient circumstances. Always consult with a qualified healthcare professional for guidance. See additional information.